[Full PDF of report is available below]
Modeling in epidemiology can serve as a useful alternative to reality, as it is often impossible to observe and record all real interactions across highly complex systems. By attempting to reduce the system to a series of equations or probability-based distributions, it is possible to produce outcomes that might reflect, to a useful extent, what may happen under certain conditions in nature. It is far cheaper and faster than conducting an observational study of long duration across diverse epidemiological settings.
The attractiveness of turning years of massive parallel studies into a few seconds of high-powered computing is obvious. However, being completely dependent on both the design of the program and the input parameters the program is instructed to compute, the outputs of models are more akin to a picture painted by humans than a cinematic record of a natural phenomenon. Like a two-dimensional painting, it can provide a useful approximation of reality if the artist so wishes and is sufficiently skilled. Alternatively, it can provide a picture that leads the viewer to see things that don’t occur in nature, exaggerating certain aspects whilst minimizing others, which by design or by accident can elicit emotions or reactions that direct observation may not produce. While providing important insights, it is at best a rough imitation.
Modeling of human disease becomes still more complicated when it is intended to predict rare events at a population level, as the conditions and responses that promote or mitigate diseases change greatly over time. Infectious diseases previously killed about half of all children before the age of 10, but mortality is now relatively rare in wealthier countries due predominantly to changes in hygiene, living conditions, nutrition, and the advent of antibiotics. Massive mortality events such as the Black Death, probably due to the bacteria Yersinia pestis, are now extremely unlikely because environmental conditions that promoted them are less prevalent and the infection is readily treated with common antibiotics. Reliance on such historical events to predict the likelihood of current health risks would be like predicting the safety of modern air travel based on the performance of the Wright brothers’ original airplane designs.
Since early in the Covid-19 outbreak, and indeed some years before, there has been an increased international public health emphasis on the risk of outbreaks and pandemics. While this may seem incongruous in light of the overall steady global reduction in infectious disease mortality over the past 30 years, the concern has led to requests for unprecedented funding and a major reorientation of several international health agencies. A report published in 2024 by the REPPARE project at the University of Leeds, Rational Policy Over Panic, demonstrated that the risk had been misrepresented in the reports of several key international agencies involved in pandemic prevention, preparedness, and response (PPPR) policy development. A significant reason was a failure to consider advances in health care and technological advances to detect and record disease outbreaks.
With the acute phase of the Covid-19 pandemic over, many countries are reviewing their public health response and the priority and manner with which future pandemic risk should be addressed. Member States of the World Health Organization continue discussions on the proposed Pandemic Agreement and acceptance of recent amendments to the International Health Regulations. Contemporaneously, several new PPPR institutions have already been established, including a new Pandemic Fund, International Pathogen Surveillance Network, and a Medical Countermeasures Platform, all of which are updating their investment cases and financial requirements.
Predictive modeling by Metabiota, a company now absorbed by Ginkgo Bioworks, has contributed significantly to the conversation on pandemic risk and the need for increased financing. It constituted one of two main sources for assessment of risk in the G20 High Level Independent Panel (HLIP) report in June 2021, which was influential in informing the G20 Group of Nations’ support for the WHO’s PPPR agenda. REPPARE previously addressed concerns regarding the interpretation of model outputs based on a paper by Meadows et al. (2023) which included Metabiota (Ginkgo Bioworks) authorship. Ginkgo Bioworks have now provided a more detailed report to the New Zealand Royal Commission on COVID-19 Lessons Learned – Estimated Future Mortality from Pathogens of Epidemic and Pandemic Potential – hereafter called the Bioworks report.
The Bioworks report aims to predict the threat of epidemics and pandemics to human health. Risk is estimated through computational epidemiology and extreme events modeling simulations to estimate mortality from “low frequency, high severity” epidemics and pandemics from respiratory diseases, particularly pandemic influenza, novel coronaviruses, and viral hemorrhagic fevers (VHFs).
The relative frequency and size of predicted outbreaks can be seen in the graphic below from the Bioworks report. While nearly all events are of relatively low mortality, as all modern pandemics of confirmed natural origin have been, the main driver of average annualized ‘expected’ deaths is derived from rare but massive events of a size the world has not seen since the development of modern medicine.
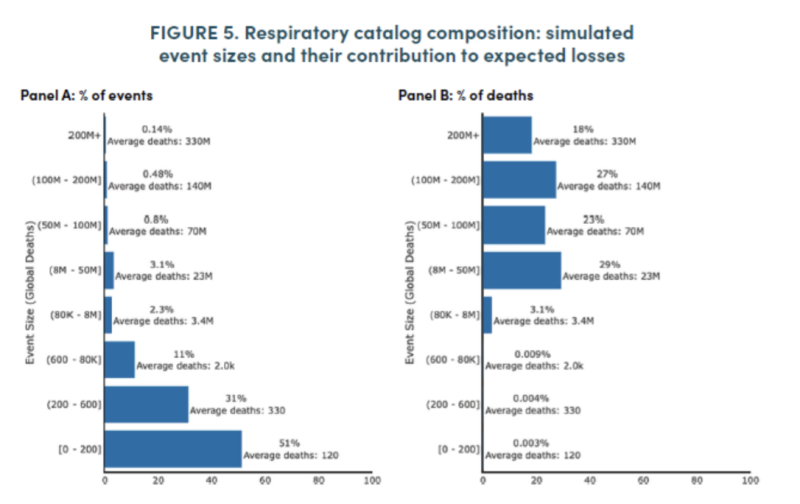

The Bioworks report concludes that an average of 2.5 million deaths are attributable annually to these acute respiratory outbreaks (1.6 million for pandemic influenza alone). Many will find these results implausible. There has not been such an annual influenza mortality in a century, and only twice in the past century, in 1957-8 and 1968-9, did the mortality rate reach what the model suggests is average. The WHO considers Covid-19, if included as a natural outbreak, has a reported mortality of just over seven million over three years.
For VHF, the report estimates an average of 26,000 globally, and 19,000 in sub-Saharan Africa. This is higher than previously recorded in any year. The largest in recent history, the 2014 Ebola outbreak, caused just 11,325 deaths. Hemorrhagic fever is predicted to exceed 100,000 every 25 years mortality with a likelihood of 48%, an event that may not have occurred in human history.
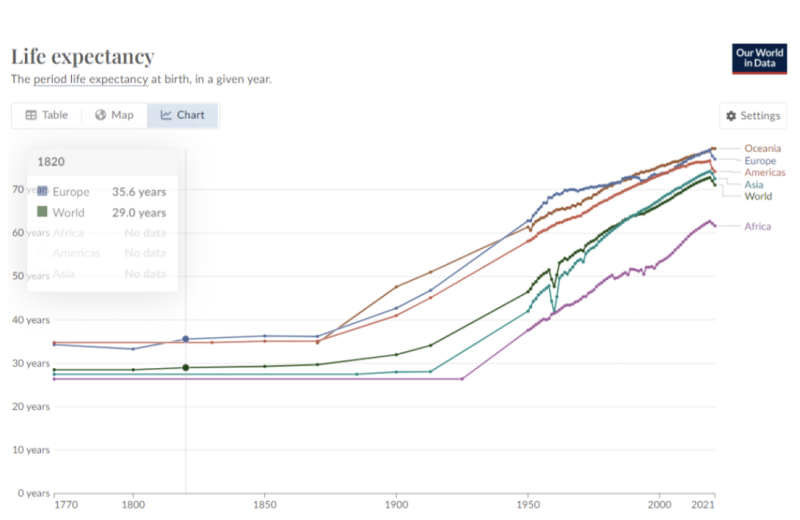
Two major oversights lead to these outcomes. First, the model overlooks changes in society and medicine over the past several hundred years that have seen average global life expectancy rise from below 30 years to over 70, and over 80 years in some wealthier countries (see below). Thus, bacterial infections such as the Plague (Y. pestis), and diseases such as cholera and typhus associated with poor hygiene are assumed to have a recurrence rate and magnitude relevant to massive historic outbreaks. Spanish influenza in 1918-19 resulted in considerable mortality due to secondary bacterial infections, which are far less likely to recur since the advent of modern antibiotics.

Second, the model fails to account for the advent of modern diagnostics such as PCR, point-of-care antigen and serology testing and genetic sequencing, and the improved ability to record and transmit such information. Thus, it is assumed that the increase in reporting reflects a real increase in outbreak frequency rather than largely reflecting improved ability to detect. The model then assumes a continuation of this increase in future years.
In view of the enormous changes in medicine over the last 100 years, and the continuing steady decrease in infectious disease mortality, the assumptions underlying the model’s predictions seem implausible. While future advances in medicine are difficult to measure, it seems reasonable to assume that the advances in hygiene practices, nutrition, housing, diagnostics, antibiotics, and vaccines over the past century will continue with further mitigation of risk in future years. While antimicrobial resistance can occur, it is a problem predominantly for endemic infections more than epidemic, and advances in antimicrobial countermeasures will continue.
Modeling of this type has become highly influential in policy development. As computing power increases, it has been tempting to think that predictive accuracy increases. However, a model with unrealistic assumptions and input parameters simply gets to an implausible outcome in a shorter period.
As an academic exercise, modeling can assist in raising questions to be answered by serious research. Yet, when misapplied and overemphasized as a guide for policy, it risks diverting financial and human resources from real disease burdens to spurious ones. This will result in increased mortality, as outcomes of current high-burden endemic infectious diseases, such as malaria and tuberculosis, remain highly dependent on the availability of official development assistance (ODA, or ‘foreign aid’). ODA for nutritional support, fundamental to improving health outcomes, has dropped 20% over the past four years. Based on predictions including the one discussed here, the equivalent of nearly 50% of pre-Covid ODA is proposed for pandemic preparation and response. This will reduce essential interventions elsewhere.
Technological advances have contributed to the reduction in infectious diseases, including pandemic mortality. A misuse of technology through inappropriate use of models could undo many of these important gains. By analogy, we don’t judge the probability of surviving trans-Atlantic air travel based on the probability of canvas wing covers ripping. Nor should we assess the probability of surviving future pandemics based on the era of Medieval medicine.
Notes:
Full report can be found at: https://essl.leeds.ac.uk/downloads/download/254/when-models-and-reality-clash-a-review-of-predictions-of-epidemic-and-pandemic-mortality
REPPARE reports on pandemic risk and financing for the pandemic preparedness and response agenda are at: https://essl.leeds.ac.uk/directories0/dir-record/research-projects/1260/reevaluating-the-pandemic-preparedness-and-response-agenda-reppare
Join the conversation:


Published under a Creative Commons Attribution 4.0 International License
For reprints, please set the canonical link back to the original Brownstone Institute Article and Author.









